R is good for logistic regression analysis through
glm function, but results are not very nice for report. Here I provide function that we call
reglog that is useful to show relevant data for the analysis of a categorical covariate.
reglog is called with a formula:
reglog(y~x+z). You will get the analysis of variable x, adjusted for z respect to the binary response y. While x and y will be treated as factors if they are nor when suplied, covariates for adjustment can be numerical or factors. You can adjust for anything allowed in a
glm formula, including interactions, but will not be shown. Interactions with x are not allowed. In a future post I will provide functions to study interactions.
Similar to glm,
reglog accepts a
data argument with a data.frame containing the variables. Missing values are excluded and a subset argument can be used to select observations for analysis.
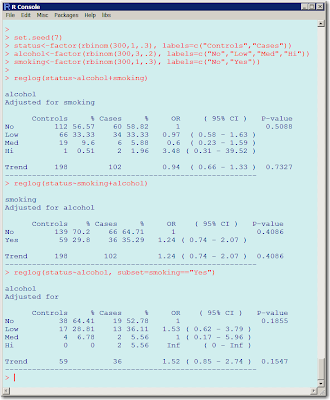
Sorry about the quality of this image. Blogspot doesn't allow better resolution and if I paste the text will not respect the spacing even if I use &nsbp;


